Clinical Trial Integrated Matching System (CTIMS)
Improve efficiencies. Accelerate Cancer Research. Match patients to clinical trials.
Clinical trials play an important role in scientific discovery, yet the process to match patients to clinical trials is highly manual, enormously time consuming, and prone to error. Nearly 25% of patients diagnosed with cancer at Princess Margaret Cancer Center are matched to clinical trials, while many more eligible patients are missed.
Prior to the CTIMS project, there were efforts to improve the effectiveness and efficiency of clinical trial matching with technology, leveraging open-source software such as MatchMiner (from the Dana Farber Cancer Institute), and cBioPortal. Even with these tools, clinical trial matching was still a very manual and time-consuming process, sometimes taking up to 4 weeks to match patients to a clinical trial. The purpose of the CTIMS Grand Challenge project was to optimize established workflows and build new open-source software to automate high-quality clinical trial matching.
By refining existing processes and introducing innovative tools, CTIMS offers a new trial match process that significantly minimizes the time and resources required to identify patients eligible for clinical trials.
Trial Matchi Workflow: Making clinical trials matching easier
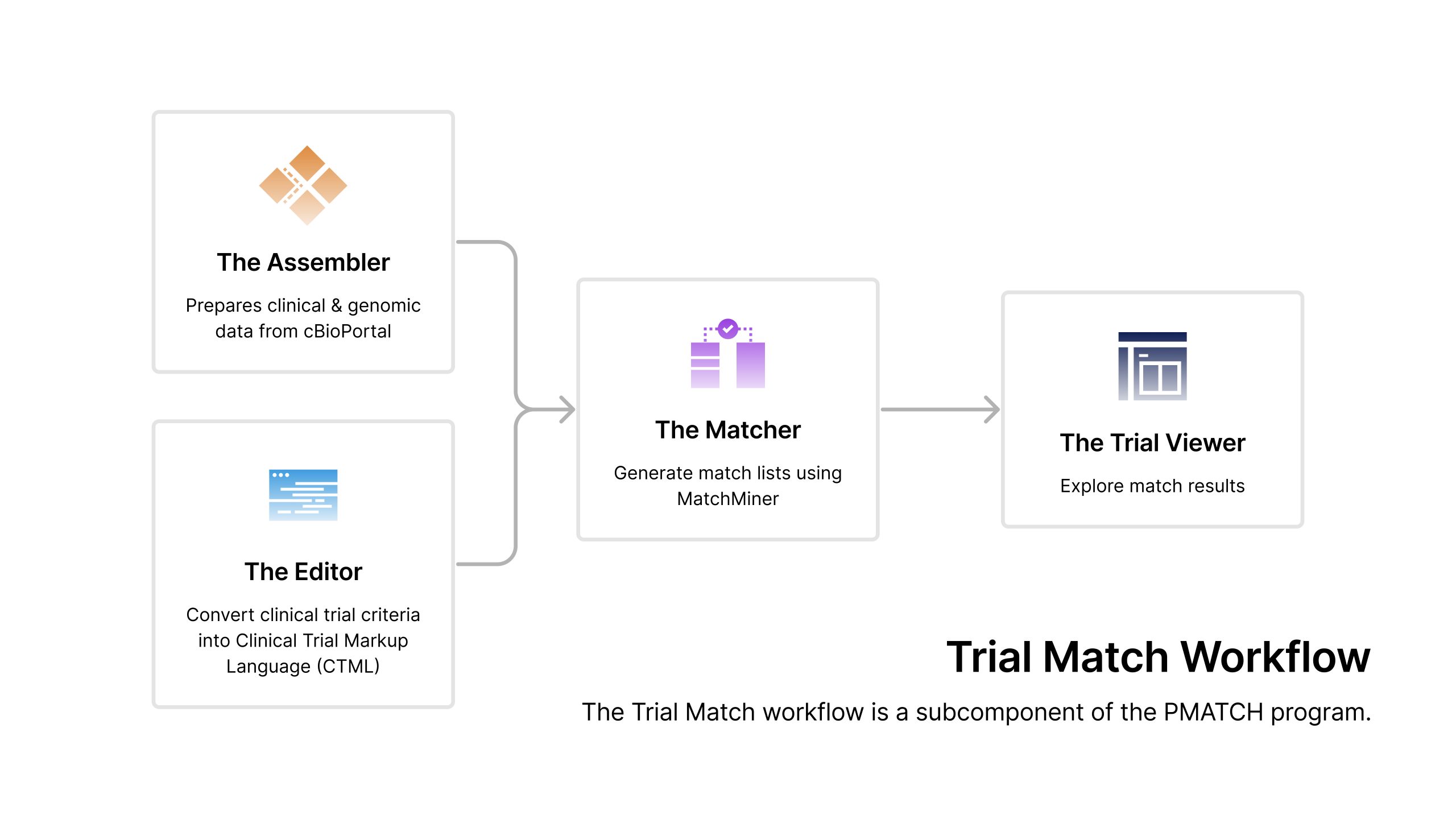
The Assembler
Improvements to the existing Assembler further enable the aggregation and transformation of de-identified patient data (clinical and genomic) for matching purposes. Once clinical and genomic data are assembled, files are sent to the matcher.
The Editor
This novel web-application provides a user-friendly tool for transcribing clinical trial criteria and seamlessly translating them into a format (Clinical Trial Markup Language or CTML) suitable for matching. Once a CTML is created, file is sent to the matcher.
In comparison to older processes, the Editor streamlines operations by automating over half of the manual tasks. Simultaneously, it reduces the number of free text fields by standardizing inputs, serving as a safeguard against misspelling and inconsistencies. These improvements are crucial for reliably matching patients with suitable clinical trials.
Features:
- Intuitive, user-friendly form to easily capture eligibility criteria in accordance with clinical trial protocols.
- Accurately reflect intricate trial inclusion/exclusion criteria for each trial arm.
- Verify CTML formatting for each trial arm with the aid of a preview window.
- Efficiently oversee and collaborate on all CTMLs within a trials team.
- Automatically convert clinical trial details into CTML format that can be exported or sent to The Matcher with the click of a button.
The Matcher
Improvements to the existing Matcher expands matching capabilities to align with the ever-growing complexities of clinical trials and introduces automation to seamlessly load CTMLs for matching.
Comprehensive validation was conducted using manually curated trial-match data from 12 completed clinical trials to assess performance across real-world patient and eligibility criteria. CTIMS analysis and validation will continue, but the current system exhibits robust matching capabilities.
The Viewer
A new way for viewing patient details matched to clinical trials.
What’s next?
The success of CTIMS was leveraged to secure funding from Genome Canada and forge exciting collaborations with esteemed institutions like Dana Farber Cancer Institute and Memorial Sloan Kettering Cancer Center. Together, we are committed to enhancing clinical trial matching processes across institutions.



 PMATCH
PMATCH
Improvements of the CTIMS project will transition to the Princess Margaret Patient Matching (PMATCH) initiative. PMATCH aims to increase clinical trial efficiency and matchability by interlinking various important data systems required for systematic, data-driven clinical decision-making.
Piloting with Active Clinical Trials
To further enhance the Trial Matching workflow, we will leverage data from 50 new trials to pinpoint areas for improvement and establish a more robust and inclusive process for matching patients with clinical trials.
Additional Information
CTIMS is the winner of the 2022-23 Princess Margaret Grand Challenge. The Grand Challenge competition offers support from the Cancer Digital Intelligence (CDI) program in front-end and back-end development, data science, design, and project management. Winning projects are selected based on their alignment with CDI priorities, contribution to the PM community, feasibility, and impact. The next Grand Challenge open call is coming in 2023.
This project is a collaboration between Dr. Trevor Pugh’s lab and members of the CDI program. Team members: Trevor Pugh, Prasanna Kumar Jagannathan, Marian Tang, Benjamin Haibe-Kains, Kelly Lane, Tran Truong, Sharon Narine, Benjamin Grant, Anton Sukhovatkin, Mickey Ng, Pietro Andreoli, Srimathi Jayasimman, Adam Badzynski, Philippe Bedard and Sophie Cooke.
Contact Information
To learn more about the Pugh Lab, visit them at https://pughlab.org/.
To learn more about CTIMS, click here.
To contact the team, email cbioportal_group@uhnresearch.ca.