An AI-Driven Intraoperative Diagnostic Tool for Detecting Cancer
Transforming surgical decision-making with AI
The Problem: Delays to diagnosing central nervous system (CNS) cancers
Central Nervous System (CNS) cancer impacts thousands of Canadians each year, leading to poor health outcomes and low survival rates. Current neurosurgical diagnostic methods involve intraoperative consultation with a neuropathologist and post-operative analyses. While intraoperative consultation provides real-time insights, it is often subjective and less accurate than post-surgical methods. This may lead to suboptimal resection and the need for additional surgeries. Furthermore, post-operative analysis is time-consuming, which further delays a targeted treatment plan.
To address this, a novel intraoperative diagnostic method using picosecond infrared laser mass spectrometry (PIRL-MS) has been developed by researchers at UHN in partnership with Unity Health. This technique creates ‘fingerprints’ of tissues that have been altered by cancer in just 10 seconds. This is then compared to a library of tumor types to quickly identify the cancer type.
The Solution: Leveraging AI to enhance diagnostic accuracy
This Grand Challenge project aims to leverage machine learning predictions, with PIRL-MS to enhance the classification of CNS cancer, through the development of an intra-operative application. Designed for pathologists in the laboratory, the machine learning predictions will be used to facilitate decision-making by providing real-time and accurate diagnostic classifications to reduce the time and resources required and to ultimately optimize patient outcomes.
Project Goal
The goal of this project is to develop a software application that provides machine learning-powered decision support for pathologists to quickly and accurately classify high-priority groups of CNS cancers in the laboratory setting.
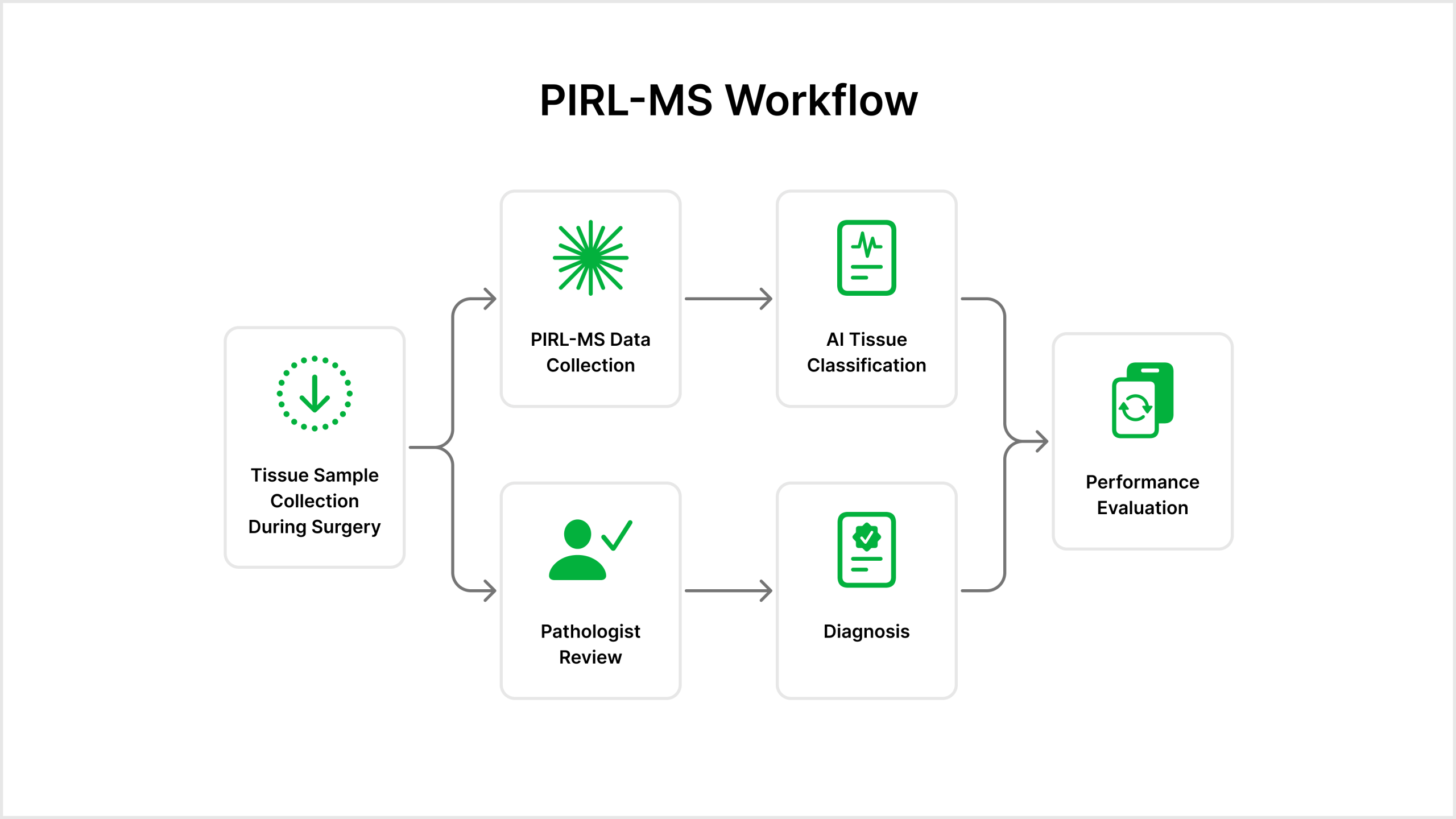
The team aims to develop a medical application for intraoperative use as a decision support tool in CNS cancer surgeries. The application will feature:
- A user-friendly pathologist interface
- A streamlined interface for entering data on de-identified patients, optimized for use during surgery
- Integration with PIRL-MS device
- This tool will be built with the goal of connecting to the PIRL-MS device to collect tissue samples in real
- Automated Data Pipeline
- Tissue samples will be processed through a pipeline that:
- Captures laser tissue data via PIRL-MS
- Performs quality control and pre-processing
- Outputs a CNS cancer classification based on the mass spectrometry data
- Tissue samples will be processed through a pipeline that:
Ultimately, the application will be designed to support rapid, accurate decision making for pathologists, while also enabling data collection for future model refinement
Project Deliverables
Within a 1-year timeline, the CDI project team will work towards completing the following deliverables:
- Responsible AI Framework Support: Apply the framework to provide feedback and improve on
- Develop Application: Create a user-friendly decision support tool to surface model classification details
- This application aims to enhance accuracy for identifying high-priority groups of CNS cancer types using Picosecond Infrared Laser Mass Spectrometry (PIRL-MS).
Additional Information
This project marks the launch of an exciting partnership between the Cancer Digital Intelligence program, at the Princess Margaret Cancer Centre, with the University of Waterloo. By combining Waterloo’s cutting-edge research, and UHN’s clinical excellence, this partnership creates a powerful pathway to accelerate the translation of innovative AI solutions into clinical settings.
Read more about the exciting partnership here.
This project is the winner of the 2024-2025 Princess Margaret Grand Challenge and is the first collaboration with the University of Waterloo. The Grand Challenge offers support from the Cancer Digital Intelligence (CDI) program in the form of front-end and back-end development of the project, data science, design, and project management support. Winning projects are selected based on their responsible use of AI, or addressing risks of AI adoption in their projects, alignment with CDI values and priorities, contribution to the PM community, and feasibility of completion within the Grand Challenge funding period, and impact.
This project is a collaboration between Dr. Scott Hopkins, Dr. Arash Zarrine-Afasr, and members of the CDI team. Team members: Dr. Scott Hopkins, Dr. Arash Zarrine-Afsar, Kelly Lane, Tran Truong, Benjamin Grant, Helena Hyams, Anton Sukhovatkin, Pietro Andreoli, Adam Badzynski, Darah Vlaminck, Francis Talbot, Cailum Stienstra, Patrick Thomas, Alexa Fiorante.
Dr. Scott Hopkins is Professor of Chemistry at the University of Waterloo. His research interest lies in the intersection of chemistry and physics, receiving numerous awards for his research over the years.
To learn more about Dr. Scott Hopkins, and his work, click here:
https://uwaterloo.ca/chemistry/profile/shopkins
Dr. Arash Zarrine-Afsar is trained as a physical biochemist. He is currently serving as a Principal Investigator at UHN and leads groups of researchers focused on the development of rapid diagnostic methods using mass spectrometry.
To learn more about Dr. Arash Zarrine Afsar and his work, click here:
https://www.uhnresearch.ca/researcher/arash-zarrine-afsar
https://medbio.utoronto.ca/faculty/zarrine-afsar